Biostasis and Intrisically Disordered Proteins
Harvard, 2020 - ?
This is my current main project as a Postdoctoral Associate in the Marks Lab, in the Systems Biology department at Harvard Medical School. In direct continuation of my goal to understand the emergence of living systems' properties at different scales, this research represents a deep dive into molecular biology and bioinformatics, with a specific focus on the "disordered" proteome. This family of proteins, whose omnipresence yet resistance to many traditional means of investigations have earned it the name of Dark Proteome, seems to (at least sometimes) challenge the structure-function paradigm.
The recent years have seen an increased interest in the intrinsically disordered regions of proteins (IDP). They are involved in just about every important cell biological processes, and are recognized as drivers of "membrane-less" organelles where they form compartments of macroscopic droplets without the need of lipids - which also hints at the important role they very likely played in the apparition of life on earth. Together with the association of specific IDPs with prion-like mechanisms and amyloidosis contribution, the challenge to "understand" IDPs is quite pressing. However, in contrast to our collective knowledge of proteins that form relatively well specified subfamilies, 3D structures with known functions, functional classification of so-called disordered protein sequences lags far behind, often relying on alignments of the non-disordered regions. The main questions I am trying to address in this research revolve around finding a way to functional classification of IDP sequences.
I have been heavily involved on the computational side of the DARPA funded Biostasis project, whose broader goal is to identify and synthesisze protein sequences responsible for cryptobiosis - the ability for an organism or a cell to undergo long periods of "hibernation" when the environmental conditions are not favorable for life, and then be subsequently ressucitated. IDPs are heavily involved in most natural examples of cryptobiosis (of which Tardigrades are definitely a good ambassador). Therefore, I have been devising new ways to represent and classify disordered protein sequences, and have been trying to correlate them with cryptobiosis characteristics. Notably, I have worked on representing any sequence of Amino Acids into a spectral domain from which I extract and compare salient features using Machine Learning. This spectral representation highlights periodicity information that is relevant to many of the known characteristics of interactions between IDPs.
-
Scalable nonparametric Bayesian models that predict and generate genome sequences.
Neurips Workshop, 2021 - + journal article in preparation
Engineered Living Material
MIT, 2018 - 2020
This was my first and main research project as a Postdoctoral Associate at MIT. I was working under the direct supervision of Prof. Neri Oxman and in close collaboration with Profs. Christopher Voigt, Cullen Buie, and Timothy Lu. The ambitious goal of this project is to program and simulate the growth of a multicellular aggregate, compile the virtual genotype of the simulated organisms into real DNA, and subsequently implant this into real cells. At present, the model organism of interest is Escherichia coli.
We want to be able to computationally design a phenotype - a desired pattern - and have the software output the corresponding DNA code (genotype). This DNA will then reproduce the same growth process and the corresponding patterns when inserted into a living cell.
I have developed a specialized agent-based simulation engine, a logic-gate based virtual gene regulatory network and a machine learning framework that heavily relies on evolutionary algorithms. The machine learning part is particularly fascinating, as it applies to both the calibration of the simulation (trying to reduce the reality gap) and the inverse problem (finding the correct genotype for a desired phenotype).
-
Viva in Silico: A position-based dynamics model for microcolony morphology simulation.
Artificial Life Conference, 2018 - + journal article in preparation
MecaCell & Cell Physics Simulation
2014 - 2019
Since a considerable amount of my research required state of the art cell physics simulation, I developed a specialized cell physics engine. MecaCell is an open-source platform for the agent-based simulation of cells, with a strong focus on modularity and performance. Depending on the core physics model used, it is able to handle several millions of cells on a conventional CPU. It is a lightweight platform designed with artificial life and morphogenetic engineering in mind. It has been in use throughout most of my PhD and for my current Postdoc - and it keeps getting better.
-
MecaCell: an Open-source Efficient Cellular Physics Engine
Artificial Life Conference, 2015 -
Viva in Silico: A position-based dynamics model for microcolony morphology simulation.
Artificial Life Conference, 2018 -
Learning Aquatic Locomotion with Animats
ECAL, 2017 - MecaCell on github
Artificial Gene Regulatory Networks
University of Toulouse, 2014 - 2018
I have used and researched better representation of aGRNs both as cell controllers (for cell cycle regulation) and as controllers for more abstract problems (for example, in this video, a GRN was evolved to control a ship).
This research gave insights on the dynamics of these complex systems and allowed for interesting improvements and variations in their design, encoding, and evolvability.
-
A comparison of Genetic Regulatory Network Dynamics and Encoding
GECCO, 2017 -
Dangerousness Metric for Gene Regulated Car Driving
Evostar, 2016 -
Artificial Gene Regulatory Networks for Agent Control
Evolutionary Computation in Gene Regulatory Network Research -
Investigating Artificial Cells’ Spatial Proliferation with a Gene Regulatory Network
Procedia computer science, 2017
Genetic Algorithm Framework
University of Toulouse, 2014 - 2018 + MIT, 2018 - 2019
My research led me to use a wide variety of evolutionary computations & genetic algorithms that, in some cases, required a degree of customization and versatility that I didn't find in other libraries. I've thus put everything together in a fun modern C++ project that keeps growing.

Self Assembly of cells through differential adhesions
EPFL, 2016
This is a soft robotics research project I worked on while a visiting PhD student at EPFL in Pr. Dario Floreano's team. By letting each deformable cell unit define its adhesion pattern, unconnected cells can, through random motion, assemble into desired geometric structures, such as the formation of snowflakes patterns.
Evo-Devo Research
University Of Toulouse, 2015 - 2018
Starting from a single cell with identical local rules governing communication, growth, and specialization, a varied panel of morphologies can develop. These environmental rules and cell controllers can be discovered and explored through various evolutionary computation tools. In many of this research and experiments, virtual creatures are composed of cells housing an artificial Gene Regulatory Network (GRN). The GRN is evolved in order to favor the organisms that would best fit their environment and its energy constraints.
One of my main interest in this area is in studying how very simple fitness functions make for the emergence of complex phenotypes. For example, in a series of experiments that I did during my PhD, the organism only have to "survive"; the complex phenotypes and life-like behaviors emerge from the simulation constraints (mostly related to energy). Here, an example of growth and auto-organisation into shells (nutritive cells feeding a shield of very resistant cells), and another example showing the emergence of roots and leaves (to produce energy from simulated sunlight and nutrients buried in the ground).
-
Self-organization of symbiotic multicellular structures
Artificial Life Conference, 2014 -
Evolved developmental strategies of artificial multicellular organisms
Artificial Life Conference, 2016

Robotics
French Robotics Cup team in Toulouse, 2015 - 2019
This is more of a hobby than a "true" research project, but, to me, being a part of such a great robotics team was an invaluable opportunity to apply and improve my electronics skills, as well as some computer vision and planning algorithms.
I have designed, from scratch, the electronics of the two robots of our team that had to compete in the french robotics cup. I also wrote some low-level code and helped with computer vision algorithms. Overall, this was an incredible experience, especially since we have had excellent results (winners of the 2021 edition, Innovation Price in 2016, First Runner-Up in 2017 and 2019, 3rd at the European Robotics Cup of 2017).
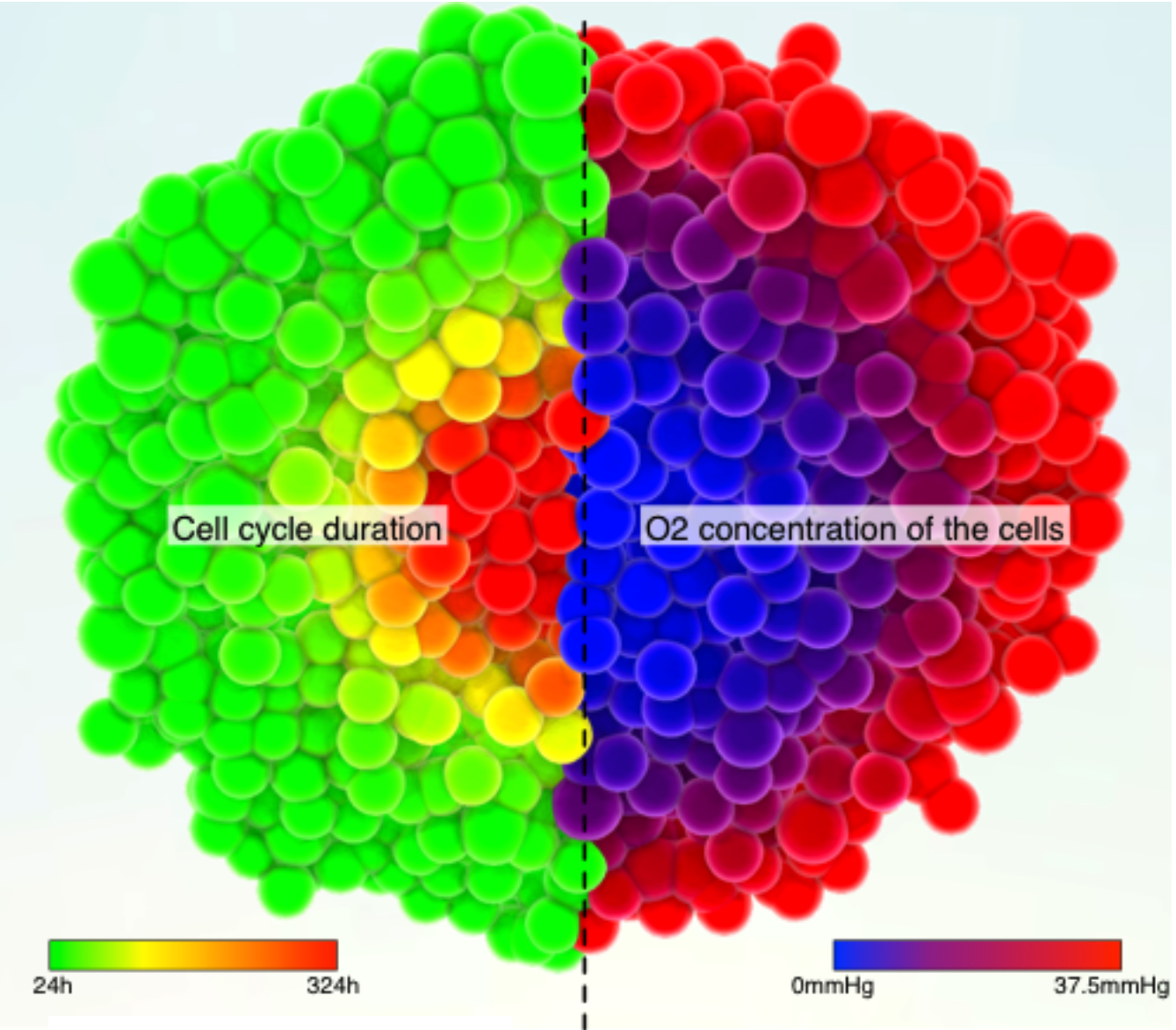
Applications to Cancer simulation + Evolutionnary dynamics of the immune system
University of Toulouse + ITAV & INSERM - 2015 - 2016
My cell simulation software has been used to model tumor spheroids and study the formation of a necrotic core due to various constraints (oxygen & nutrient depletion and mechanical constraints mostly). Combining this software with evolutionry computations, I have also helped with the simulation of the evolutionary dynamics of the "arms race" between the immune system and melanoma.